2024
37. “Recent developments in enzymatic and microbial biosynthesis of flavor and fragrance molecules.”
Dickey, R.M.; Gopal, M.R.; Nain, P.; Kunjapur, A.M.*
J. Biotechnol. In press.
(Invited contribution)
36. “Creating next-generation live vaccines by autonomous production of nitrated antigens.”
Kunjapur, A.M.*
Science. 2024 384 (6691), 41-42.
(Prize essay, not peer-reviewed)
35. “Harnessing biocatalysis to achieve selective functional group interconversion of monomers.”
Gopal, M.R.; Kunjapur, A.M.*
Curr. Opin. in Biotechnol. 2024 86, 103093.
(Invited contribution)
PDF
Dickey, R.M.; Gopal, M.R.; Nain, P.; Kunjapur, A.M.*
J. Biotechnol. In press.
(Invited contribution)
36. “Creating next-generation live vaccines by autonomous production of nitrated antigens.”
Kunjapur, A.M.*
Science. 2024 384 (6691), 41-42.
(Prize essay, not peer-reviewed)
35. “Harnessing biocatalysis to achieve selective functional group interconversion of monomers.”
Gopal, M.R.; Kunjapur, A.M.*
Curr. Opin. in Biotechnol. 2024 86, 103093.
(Invited contribution)
34. “The US Bioeconomy: Shaping our Future through Safety, Security, Sustainability, and Social Responsibility Considerations.”
Attal-Juncqua, A.; Dods, G.; Crain, N.; Dodds, D.; Evans, S.; Fackler, N.; Flyangolts, K.; Kosal, M.E.; Kunjapur, A.M.; Read, R.; Renda, B.; Scown, C.; Tchedre, K.; Ternus, K.; Vitalis, B.; Gronvall, G.K.*
Trends Biotechnol. 2023.
DOI: 10.1016/j.tibtech.2023.11.015
Open Access
33. "Reductive amination cascades in cell-free and resting whole cell formats for valorization of lignin deconstruction products"
Nain, P.; Dickey, R.M.; Somasundaram, V.; Sulzbach, M.; Kunjapur, A.M.*
Biotech & Bioeng. 2024 121 (2), 593-604.
DOI: 10.1002/bit.28604
PDF
Attal-Juncqua, A.; Dods, G.; Crain, N.; Dodds, D.; Evans, S.; Fackler, N.; Flyangolts, K.; Kosal, M.E.; Kunjapur, A.M.; Read, R.; Renda, B.; Scown, C.; Tchedre, K.; Ternus, K.; Vitalis, B.; Gronvall, G.K.*
Trends Biotechnol. 2023.
DOI: 10.1016/j.tibtech.2023.11.015
Open Access
33. "Reductive amination cascades in cell-free and resting whole cell formats for valorization of lignin deconstruction products"
Nain, P.; Dickey, R.M.; Somasundaram, V.; Sulzbach, M.; Kunjapur, A.M.*
Biotech & Bioeng. 2024 121 (2), 593-604.
DOI: 10.1002/bit.28604
32. "Selective and site-specific incorporation of non-standard amino acids within proteins for therapeutic applications."
Butler, N.D.; Kunjapur, A.M.*
Methods Mol. Biol. 2024 2720, 35-53.
(Invited contribution)
PDF
Butler, N.D.; Kunjapur, A.M.*
Methods Mol. Biol. 2024 2720, 35-53.
(Invited contribution)
2023
31. “Nitro Biosciences: Enhancing immune response via an expanded genetic code.”
Butler, N.D.*; Kunjapur, A.M.
Delaware J. Pub. Health. 2023 9 (4), 24-25.
(Not peer-reviewed)
PDF
30. "“Discovery of L-threonine transaldolases for enhanced biosynthesis of beta hydroxylated amino acids"
Jones, M.A.; Butler, N.D.; Anderson, S.; Wirt, S.; Govil, I.; Lyu, X.; Fang, Y.; Kunjapur, A.M.*
Commun. Biol. 2023 6, 929.
DOI: 10.1038/s42003-023-05293-0
Open Access
PDF
Butler, N.D.*; Kunjapur, A.M.
Delaware J. Pub. Health. 2023 9 (4), 24-25.
(Not peer-reviewed)
30. "“Discovery of L-threonine transaldolases for enhanced biosynthesis of beta hydroxylated amino acids"
Jones, M.A.; Butler, N.D.; Anderson, S.; Wirt, S.; Govil, I.; Lyu, X.; Fang, Y.; Kunjapur, A.M.*
Commun. Biol. 2023 6, 929.
DOI: 10.1038/s42003-023-05293-0
Open Access
29. "Genome engineering allows selective conversions of terephthalaldehyde to multiple valorized products in bacterial cells."
Dickey, R.M.; Butler, N.D.; Jones, M.A.; Govil, I.; Kunjapur, A.M.*
AIChE J. 2023 69 (12): e18230.
DOI: 10.1002/aic.18230
(Invited contribution to the 2023 Futures Issue)
PDF
Dickey, R.M.; Butler, N.D.; Jones, M.A.; Govil, I.; Kunjapur, A.M.*
AIChE J. 2023 69 (12): e18230.
DOI: 10.1002/aic.18230
(Invited contribution to the 2023 Futures Issue)
28. “A platform for distributed production of synthetic nitrated proteins in live bacteria.”
Butler, N.D.; Sen, S.; Brown, L.B.; Lin, M.; Kunjapur, A.M.*
Nat. Chem. Biol. 2023 19, 911-920.
DOI: 10.1038/s41589-023-01338-x
PDF
Butler, N.D.; Sen, S.; Brown, L.B.; Lin, M.; Kunjapur, A.M.*
Nat. Chem. Biol. 2023 19, 911-920.
DOI: 10.1038/s41589-023-01338-x
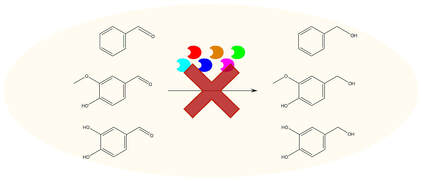
27. “Combinatorial gene inactivation of aldehyde dehydrogenases mitigates the oxidation catalyzed by E. coli resting cells.”
Butler, N.D.; Anderson, S.; Dickey, R.M; Nain, P.; Kunjapur, A.M.*
Metab. Eng. 2023 77, 294-305.
DOI: 10.1016/j.ymben.2023.04.014
PDF
Butler, N.D.; Anderson, S.; Dickey, R.M; Nain, P.; Kunjapur, A.M.*
Metab. Eng. 2023 77, 294-305.
DOI: 10.1016/j.ymben.2023.04.014
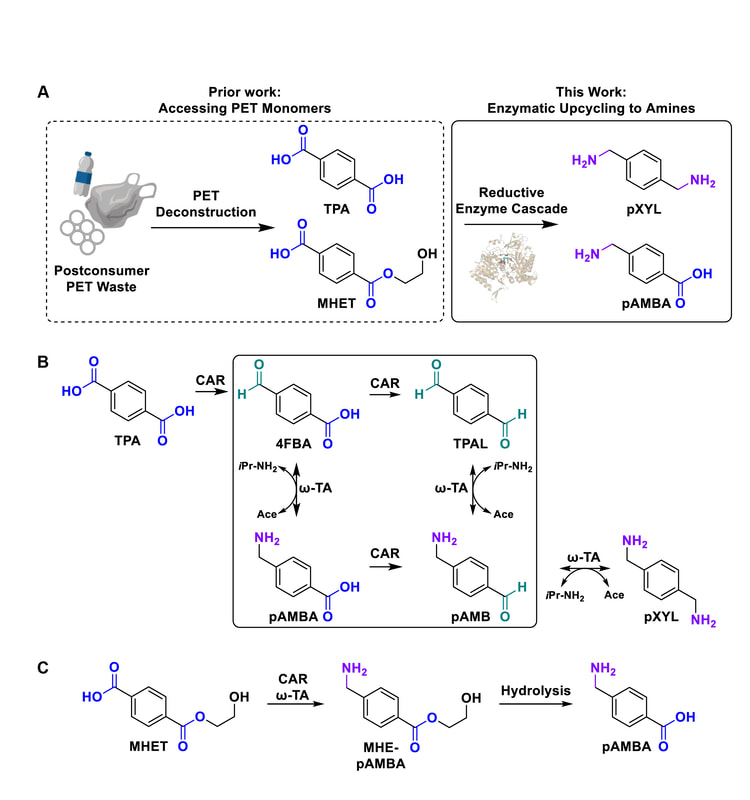
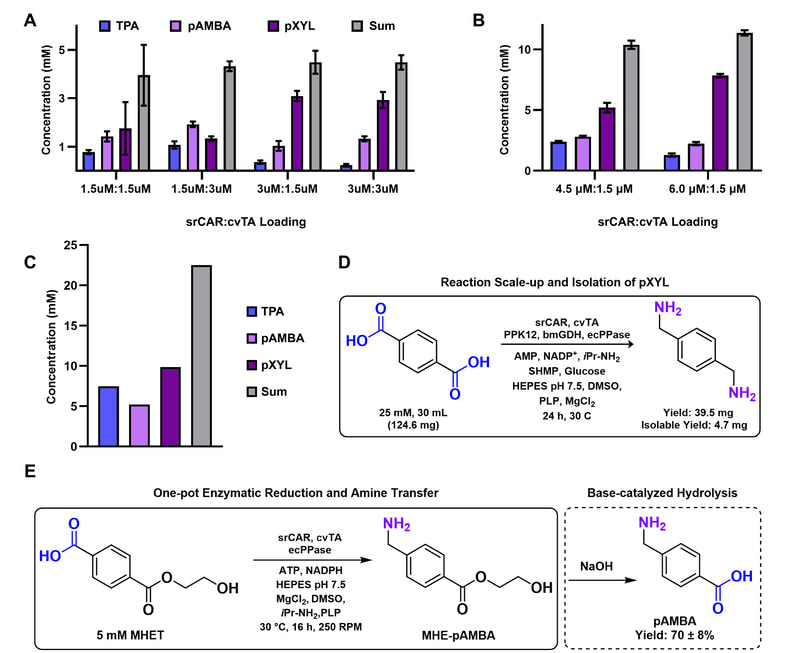
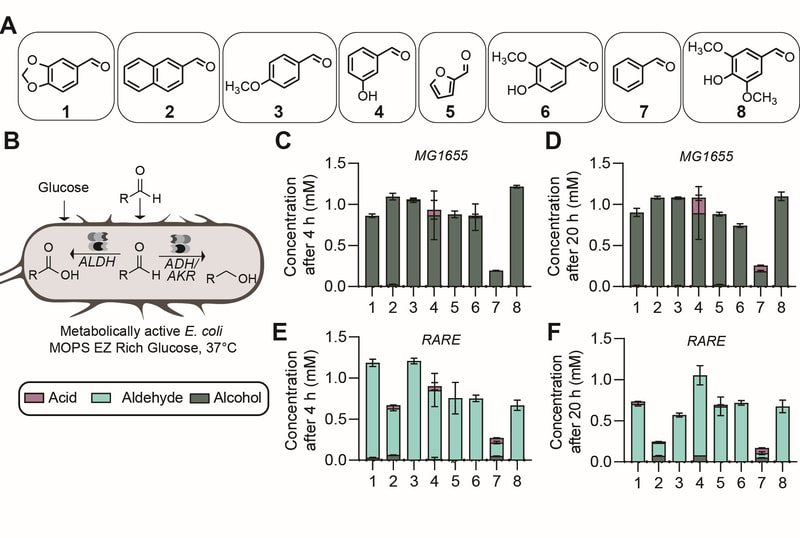
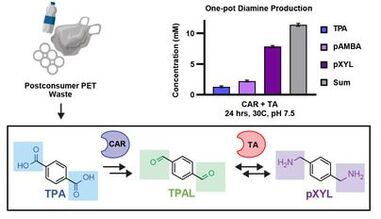
26. “Reductive enzyme cascades for valorization of PET deconstruction products.”
Gopal, M.R.; Dickey, R.M.; Butler, N.D.; Talley, M.; Nakamura, D.; Mohapatra, A.; Watson, M.P.; Chen, W.; Kunjapur, A.M.*
ACS Catal. 2023 13 (7), 4778–4789.
DOI: 10.1021/acscatal.2c06219
PDF
Gopal, M.R.; Dickey, R.M.; Butler, N.D.; Talley, M.; Nakamura, D.; Mohapatra, A.; Watson, M.P.; Chen, W.; Kunjapur, A.M.*
ACS Catal. 2023 13 (7), 4778–4789.
DOI: 10.1021/acscatal.2c06219
25. “Coordinated microbial lysis bursts into the drug delivery scene.”
Sen, S.; Kunjapur, A.M.*
Trends Biotechnol. 2023 41 (3), 295-297.
DOI: 10.1016/j.tibtech.2023.01.010
(Invited Spotlight article, not peer-reviewed)
PDF
Sen, S.; Kunjapur, A.M.*
Trends Biotechnol. 2023 41 (3), 295-297.
DOI: 10.1016/j.tibtech.2023.01.010
(Invited Spotlight article, not peer-reviewed)
2022
24. "MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters."
Terlouw, B.R.; Blin, K.; (total of 80 co-authors including Kunjapur, A.M.); Weber, T.; Medema, M.H.
Nucleic Acids Res. 2022 51 (D1), D603-D610.
DOI: 10.1093/nar/gkac1049
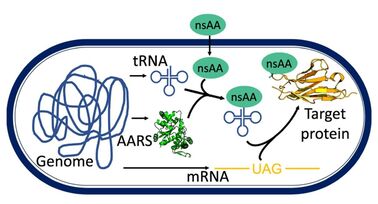
23. "Incorporation of a Chemically Diverse Set of Non-Standard Amino Acids into a Gram-Positive Organism."
Stork, D.A.; Jones, M.A.; Garner, E.C.*; Kunjapur, A.M.*
Bio-protocol. 2022 12 (17), e4507-e4507.
DOI: 10.21769/BioProtoc.4507
22. “Innovations toward the valorization of waste plastics.”
Hinton, Z.R.; Talley, M.R.; Kots, P.A.; Le, A.V.; Zhang, T.; Mackay, M.E.; Kunjapur, A.M.; Bai, P.; Vlachos, D.G.; Watson, M.P.; Berg, M.C.; Epps, T.H.; Korley, L.T.J.
Annu. Rev. Mater. Res. 2022 52, 249-280.
DOI: 10.1146/annurev-matsci-081320-032344
Terlouw, B.R.; Blin, K.; (total of 80 co-authors including Kunjapur, A.M.); Weber, T.; Medema, M.H.
Nucleic Acids Res. 2022 51 (D1), D603-D610.
DOI: 10.1093/nar/gkac1049
23. "Incorporation of a Chemically Diverse Set of Non-Standard Amino Acids into a Gram-Positive Organism."
Stork, D.A.; Jones, M.A.; Garner, E.C.*; Kunjapur, A.M.*
Bio-protocol. 2022 12 (17), e4507-e4507.
DOI: 10.21769/BioProtoc.4507
22. “Innovations toward the valorization of waste plastics.”
Hinton, Z.R.; Talley, M.R.; Kots, P.A.; Le, A.V.; Zhang, T.; Mackay, M.E.; Kunjapur, A.M.; Bai, P.; Vlachos, D.G.; Watson, M.P.; Berg, M.C.; Epps, T.H.; Korley, L.T.J.
Annu. Rev. Mater. Res. 2022 52, 249-280.
DOI: 10.1146/annurev-matsci-081320-032344
2021
21. “Advances in engineering microbial biosynthesis of aromatic compounds and related compounds.”
Dickey, R.M.†; Forti, A.M.†; Kunjapur, A.M.*
Bioresour. Bioprocess. 2021 8, 91.
(Invited review)
DOI: 10.1186/s40643-021-00434-x
20. “Designing efficient genetic code expansion in Bacillus subtilis to gain biological insights.”
Stork, D.A.; Squyres, G.R.; Kuru, E.; Gromek, K.A.; Rittichier, J.; Jog, A.; Burton, B.M.; Church, G.M.*; Garner, E.C.*; Kunjapur, A.M.*
Nat. Commun. 2021 12, 5429.
DOI: 10.1038/s41467-021-25691-4
Open Access
Dickey, R.M.†; Forti, A.M.†; Kunjapur, A.M.*
Bioresour. Bioprocess. 2021 8, 91.
(Invited review)
DOI: 10.1186/s40643-021-00434-x
20. “Designing efficient genetic code expansion in Bacillus subtilis to gain biological insights.”
Stork, D.A.; Squyres, G.R.; Kuru, E.; Gromek, K.A.; Rittichier, J.; Jog, A.; Burton, B.M.; Church, G.M.*; Garner, E.C.*; Kunjapur, A.M.*
Nat. Commun. 2021 12, 5429.
DOI: 10.1038/s41467-021-25691-4
Open Access
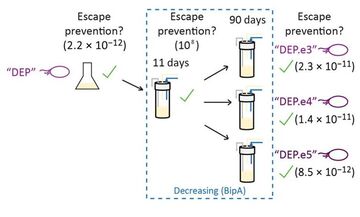
19. "Synthetic auxotrophy remains stable after continuous evolution and in co-culture with mammalian cells."
Kunjapur, A.M.†*; Napolitano, M.G.†; Hysolli, E.†; Noguera, K.; Appleton, E.M.; Schubert, M.G.; Jones, M.A.; Iyer, S.; Mandell, D.J.; Church, G.M.*
Sci. Adv. 2021 7 (27), eabf5851.
†Contributed equally.
DOI: 10.1126/sciadv.abf5851
Open Access
Kunjapur, A.M.†*; Napolitano, M.G.†; Hysolli, E.†; Noguera, K.; Appleton, E.M.; Schubert, M.G.; Jones, M.A.; Iyer, S.; Mandell, D.J.; Church, G.M.*
Sci. Adv. 2021 7 (27), eabf5851.
†Contributed equally.
DOI: 10.1126/sciadv.abf5851
Open Access
18. "Recombineering and MAGE."
Wannier, T.M.; Ciaccia, P.; Ellington, A.; Filsinger, G.; Isaacs, F.; Javanmardi, K.; Jones, M.A.; Kunjapur, A.M.; Nyerges, A.; Pal, C.; Church, G.M.
Nat. Rev. Methods Primers. 2021 1 (7).
DOI: 10.1038/s43586-020-00006-x
Wannier, T.M.; Ciaccia, P.; Ellington, A.; Filsinger, G.; Isaacs, F.; Javanmardi, K.; Jones, M.A.; Kunjapur, A.M.; Nyerges, A.; Pal, C.; Church, G.M.
Nat. Rev. Methods Primers. 2021 1 (7).
DOI: 10.1038/s43586-020-00006-x
2020
17. “Deployment of engineered microbes: Contributions to the bioeconomy and considerations for biosecurity.”
Parker, M.; Kunjapur, A.M.*
Health Secur. 2020 18 (4), 278-296.
DOI: 10.1089/hs.2020.0010
16. “The pathway less traveled: Engineering biosynthesis of nonstandard functional groups.”
Sulzbach, M.; Kunjapur, A.M.*
Trends Biotechnol. 2020 38 (5), 532-545.
DOI: 10.1016/j.tibtech.2019.12.014
15. “Carboxylic acid reductases in metabolic engineering.”
Butler, N.D.; Kunjapur, A.M.*
J. Biotechnol. 2020 307 (1), 1-14.
DOI: 10.1016/j.jbiotec.2019.10.002
(First paper from the Kunjapur Lab at U. Delaware)
Parker, M.; Kunjapur, A.M.*
Health Secur. 2020 18 (4), 278-296.
DOI: 10.1089/hs.2020.0010
16. “The pathway less traveled: Engineering biosynthesis of nonstandard functional groups.”
Sulzbach, M.; Kunjapur, A.M.*
Trends Biotechnol. 2020 38 (5), 532-545.
DOI: 10.1016/j.tibtech.2019.12.014
15. “Carboxylic acid reductases in metabolic engineering.”
Butler, N.D.; Kunjapur, A.M.*
J. Biotechnol. 2020 307 (1), 1-14.
DOI: 10.1016/j.jbiotec.2019.10.002
(First paper from the Kunjapur Lab at U. Delaware)
2019
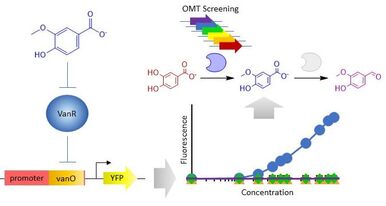
14. "Development of a vanillate biosensor for the vanillin biosynthesis pathway in E. coli.”
Kunjapur, A.M.*; Prather, K.L.J.*
ACS Synth. Biol. 2019 8 (9), 1958-1967.
DOI: 10.1021/acssynbio.9b00071
Kunjapur, A.M.*; Prather, K.L.J.*
ACS Synth. Biol. 2019 8 (9), 1958-1967.
DOI: 10.1021/acssynbio.9b00071
13. "EVOLTHON: A community endeavor to evolve lab evolution."
Strauss, S.K.; Schirman, D.; Jona, G.; Brooks, A.N.; Kunjapur, A.M.; et al.
PLoS Biol. 17 (3): e3000182.
DOI: 10.1371/journal.pbio.3000182
Strauss, S.K.; Schirman, D.; Jona, G.; Brooks, A.N.; Kunjapur, A.M.; et al.
PLoS Biol. 17 (3): e3000182.
DOI: 10.1371/journal.pbio.3000182
2018
12. “Gene synthesis allows biologists to source genes from farther away in the tree of life.”
Kunjapur, A.M.†*; Pfingstag, P.†; Thompson, N.C.*
Nat. Commun. 2018 9, 4425.
†Contributed equally.
DOI: 10.1038/s41467-018-06798-7
11. “Adaptive evolution of genomically recoded Escherichia coli.”
Wannier, T.M.†*; Kunjapur, A.M.†*; Rice, D.P.; McDonald, M.J.; Desai, M.M.; Church, G.M.*
Proc. Natl. Acad. Sci. U.S.A. 2018 115 (12), 3090-3095.
†Contributed equally.
DOI: 10.1073/pnas.1715530115
(Highlighted by Harvard Medicine News and by PNAS)
Wannier, T.M.†*; Kunjapur, A.M.†*; Rice, D.P.; McDonald, M.J.; Desai, M.M.; Church, G.M.*
Proc. Natl. Acad. Sci. U.S.A. 2018 115 (12), 3090-3095.
†Contributed equally.
DOI: 10.1073/pnas.1715530115
(Highlighted by Harvard Medicine News and by PNAS)
10. “From designing the molecules of life to designing life: future applications derived from advances in DNA technologies.”
Kohman, R.†; Kunjapur, A.M.†; Hysolli, E.†; Wang, Y.†; Church, G.M.
Angew. Chem. Int. Ed. 2018 57 (16), 4313-4328.
†Contributed equally.
DOI: 10.1002/anie.201707976
9. “Engineering post-translational proofreading to discriminate non-standard amino acids.”
Kunjapur, A.M.*; Stork, D.A.; Kuru, E.; Vargas-Rodriguez, O.; Landon, M.M.; Söll, D.*; Church, G.M.*
Proc. Natl. Acad. Sci. U.S.A. 2018 115 (3), 619-624.
DOI: 10.1073/pnas.1715137115
(Highlighted by Harvard Medicine News)
Kohman, R.†; Kunjapur, A.M.†; Hysolli, E.†; Wang, Y.†; Church, G.M.
Angew. Chem. Int. Ed. 2018 57 (16), 4313-4328.
†Contributed equally.
DOI: 10.1002/anie.201707976
9. “Engineering post-translational proofreading to discriminate non-standard amino acids.”
Kunjapur, A.M.*; Stork, D.A.; Kuru, E.; Vargas-Rodriguez, O.; Landon, M.M.; Söll, D.*; Church, G.M.*
Proc. Natl. Acad. Sci. U.S.A. 2018 115 (3), 619-624.
DOI: 10.1073/pnas.1715137115
(Highlighted by Harvard Medicine News)
2017
8. "Material properties of the cyanobacterial reserve polymer multi-L-arginyl-poly-L-aspartate (cyanophycin)."
Khlystov, N.A.†; Chan, W.Y.†; Kunjapur, A.M.; Shi, W.; Prather, K.L.J.; Olsen, B.D.
Polymer. 2017 109 (1), 238-245.
†Contributed equally.
DOI: 10.1016/j.polymer.2016.11.058.
Khlystov, N.A.†; Chan, W.Y.†; Kunjapur, A.M.; Shi, W.; Prather, K.L.J.; Olsen, B.D.
Polymer. 2017 109 (1), 238-245.
†Contributed equally.
DOI: 10.1016/j.polymer.2016.11.058.
2016
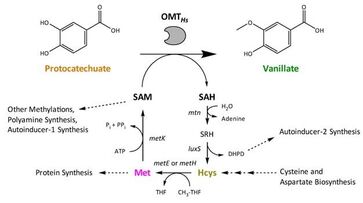
7. “Deregulation of S-adenosylmethionine biosynthesis and regeneration improves methylation in E. coli de novo vanillin biosynthesis pathway.”
Kunjapur, A.M.; Hyun, J.C.; Prather, K.L.J.
Microb. Cell Fact. 2016 15 (1), 1.
DOI: 10.1186/s12934-016-0459-x.
Kunjapur, A.M.; Hyun, J.C.; Prather, K.L.J.
Microb. Cell Fact. 2016 15 (1), 1.
DOI: 10.1186/s12934-016-0459-x.
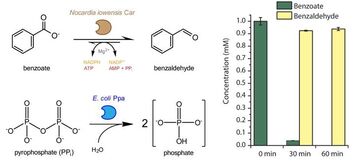
6. “Coupling carboxylic acid reductase to inorganic pyrophosphatase enhances cell-free in vitro aldehyde biosynthesis.”
Kunjapur, A.M.; Cervantes, B.; Prather, K.L.J.
Biochem. Eng. J. 2016 109 (5), 19-27.
DOI: 10.1016/j.bej.2015.12.018.
Kunjapur, A.M.; Cervantes, B.; Prather, K.L.J.
Biochem. Eng. J. 2016 109 (5), 19-27.
DOI: 10.1016/j.bej.2015.12.018.
5. “Modular and selective biosynthesis of gasoline-range alkanes.”
Sheppard, M.J.†; Kunjapur, A.M.†; Prather, K.L.J.
Metab. Eng. 2016 33, 28-40.
†Contributed equally.
DOI: 10.1016/j.ymben.2015.10.010.
Sheppard, M.J.†; Kunjapur, A.M.†; Prather, K.L.J.
Metab. Eng. 2016 33, 28-40.
†Contributed equally.
DOI: 10.1016/j.ymben.2015.10.010.
2015
4. “Microbial engineering for aldehyde synthesis.”
Kunjapur, A.M.; Prather, K.L.J.
Appl. Environ. Microbiol. 2015 81 (6), 1892-1901.
DOI: 10.1128/AEM.03319-14.
Kunjapur, A.M.; Prather, K.L.J.
Appl. Environ. Microbiol. 2015 81 (6), 1892-1901.
DOI: 10.1128/AEM.03319-14.
2014
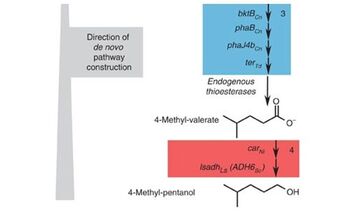
3. “Retrobiosynthetic modular approach to pathway design achieves selective pathway for microbial synthesis of the gasoline substitute 4-methyl-pentanol.”
Sheppard, M.J.; Kunjapur, A.M.; Wenck, S.J.; Prather, K.L.J.
Nat. Commun. 2014 5, 5031.
DOI: 10.1038/ncomms6031.
Sheppard, M.J.; Kunjapur, A.M.; Wenck, S.J.; Prather, K.L.J.
Nat. Commun. 2014 5, 5031.
DOI: 10.1038/ncomms6031.
2. “Synthesis and accumulation of aromatic aldehydes in an engineered strain of E. coli.”
Kunjapur, A.M.; Tarasova, Y.; Prather, K.L.J.
J. Am. Chem. Soc. 2014 136 (33), 11644-11654.
DOI: 10.1021/ja506664a.
(Highlighted by Chemical & Engineering News)
Kunjapur, A.M.; Tarasova, Y.; Prather, K.L.J.
J. Am. Chem. Soc. 2014 136 (33), 11644-11654.
DOI: 10.1021/ja506664a.
(Highlighted by Chemical & Engineering News)
2010
1. “Photobioreactor design for commercial biofuel production from microalgae.”
Kunjapur, A.M.*; Eldridge, R.B.
Ind. Eng. Chem. Res. 2010 49 (8), 3516-3526.
DOI: 10.1021/ie901459u.
Kunjapur, A.M.*; Eldridge, R.B.
Ind. Eng. Chem. Res. 2010 49 (8), 3516-3526.
DOI: 10.1021/ie901459u.